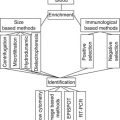
Sample
Comments
Fine-needle aspiration biopsy (FNAB), nipple aspirate with exfoliated cells
Used to determine the receptor level and metabolomic profiling [98]
Plasma
Serum
Tumor tissue
Proteomic biomarkers were isolated by laser capture microdissection of breast cancer tissue [80, 111]; a tissue microarray was used to identify candidate proteomic markers [81]; a reverse phase protein array was used to identify breast cancer biomarkers [82]; imaging biomarkers in the breast [91–95]
Epigenomic biomarkers have enormous potential and clinical implications for cancer diagnosis and prognosis. Because of the availability of genome-wide methylation, histone, and miR analysis technologies—and our rapidly accumulating knowledge regarding the epigenome—the translation of findings discussed in this chapter may be possible in the near future. Epigenetic biomarkers also may be useful in identifying patients who will benefit from a therapy without developing a resistance to the drugs. Recently developed drugs for cancer treatment are based on specific pathways and may be useful for individuals in whom these pathways are altered. This approach can be designed for personalized medicine and precision medicine. Epigenetic biomarkers also may be useful in such approaches. Additional potential breast cancer biomarkers continue to be identified, including additional high-penetrance markers.
Breast Cancer Detection Patents
A number of patents have been issued that involve genomic, epigenomic, and proteomic biomarkers (Table 20.2). Biomarkers can be used for breast cancer diagnosis either singly or in combination. These patents were generated by investigators in the industry and academic institutes. Compared to the number of publications in the breast cancer biomarker field (Table 20.3), the number of patents is low. The reason may be that many biomarkers have not been characterized in sufficient numbers of clinical samples. It is worth noting, however, that the number of reports on genetic and imaging biomarkers exceeds those of other biomarkers, as indicated in the PubMed analysis of the field shown in Table 20.3.
Table 20.2
Patents for breast cancer diagnosis and risk assessment
Patent number | Inventor/assignee | Title and comments |
|---|---|---|
US20090311671 | Rado Laboratories Ltd., Co. Antrim (GB) | Diagnosis of risk of breast cancer Polymerase chain reaction (PCR) assay of samples from breast tissue |
US5645995 | Baylor College of Medicine, Houston, TX (USA) | Methods for diagnosing an increased risk for breast or ovarian cancer Samples used were from blood, ascites, pleural fluid, and spinal fluid Point mutations in progesterone receptor gene were assayed Sequencing, single-stranded conformation polymorphism (SSCP), heteroduplexing, and restriction mapping technologies were used |
US56838885 | Baylor College of Medicine, Houston, TX (USA) | Methods for diagnosing an increased risk for breast or ovarian cancer Progesterone receptor mutations were analyzed Antibody-based assay |
US20100285456 | Matta, Jamie (USA) Carolina, R (USA) | Method for using DNA repair capacity as a biomarker for breast cancer risk in women Measurement of DNA repair capacity of lymphocytes using the host-cell reactivation assay Blood lymphocyte biomarker to calculate the probability of having carcinoma of the breast |
US7635561 | Temple University of the Commonwealth System of Higher Education, Philadelphia, PA (USA) | Methods of diagnosing, prognosing, and treating breast cancer Methylation assay in breast cancer cells obtained by biopsy Presence of methylation region in the A, B, C, and E regions of the estrogen receptor alpha gene promoter |
US20070141582 | Li, Weiwei New York (USA) Li, Jessica New York (USA) | Method for detecting early cancer and precancer in blood or body fluid Analyzed methylation levels in a panel of genes (p16, RASSF1, APC, MGMT, GSTP1, CDH-13, MLH-1) by quantitative PCR Measured demethylation of CK7, CK20, TF-1, NKX3-1, EBV, MAT-2, PAX-1, and mammaglobin-A by quantitative PCR Assay is good for breast, lung, ovarian, colon, pancreas, liver, thyroid, and nasopharyngeal cancers |
US20100221742 | National Institute of Immunology, New Delhi (IN) | Novel cancer-associated antibodies and their use in cancer diagnosis Western analysis of serum |
US20100221723 | The Institute of Columbia University in the City of New York, NY (USA) | Early detection of cancer by methylated DNA in blood Methylation assay in DNA from blood samples (serum or plasma) from breast cancer patients and controls Methylation of RASSF1A and p16 in the promoter region |
US20090035801 | Power3 Medical Products, Inc., The Woodlands, TX (USA) | 12 protein biomarkers for diagnosis and early detection of breast cancer Diagnostic assay for differentiating breast cancer, benign disease, and normal controls Based on a statistical analysis of the concentration in blood serum of one or more of the selected 12 proteins (inter-alpha–alpha trypsin inhibitor heavy chain (H4)-related protein, immunoglobulin lambda chain protein, alpha-1 microglobulin protein, apolipoprotein A-1, apolipoprotein E, complement C4 protein, serum albumin protein, lectin P35, transferrin protein) |
US6756200 | The Johns Hopkins University School of Medicine, Baltimore, MD (USA) | Aberrantly methylated genes as a marker of breast cancer malignancy Methylation of CpG islands in the promoter region of cyclin D2 by PCR-based methods of selected amplimers DNA isolated from blood, plasma, lymph, ductal cells, ductal lavage fluid, nipple aspirate, breast tissue, lymph nodes, bone marrow, or a combination of samples |
US6835541 | The Johns Hopkins University School of Medicine, Baltimore, MD (USA) | Aberrantly methylated genes as a marker of breast cancer malignancy Methylation of CpG islands in the promoter region of cRAR-beta by PCR-based methods of selected amplimers DNA isolated from blood, plasma, lymph, ductal cells, ductal lavage fluid, nipple aspirate, breast tissue, lymph nodes, bone marrow, or a combination of samples |
US20060154245 | Rigshospitalet, Copenhagen (DK) Hvidovre Hospital, Hvidovre (DK) | Method for detecting, screening, and/or monitoring a cancer in individuals Determine TIP-1 concentration from saliva samples by enzyme-linked immunosorbent assay (ELISA) and zymography Detects early-stage breast and several other cancers |
US20090221010 | Elting, J. J. Madison, CT (USA), Carney, W. P. North Andover, MA (USA), Hamer, P. J. Reading, MA (USA) | Methods for prediction and prognosis of cancer and monitoring cancer therapy Sandwich ELISA was applied to samples taken from blood, serum, plasma, urine, saliva, semen, breast exudate, cerebrospinal fluid, tears, sputum, mucous, lymph, cytosols, ascites, pleural effusions, amniotic fluid, bladder washes, and bronchioalveolar lavage Increasing levels of VEGF-165 in patient samples taken over time to detect early recurrence or metastasis |
US7662582 | Chang Gung University, Tao-Yuan (TW) | Method of identifying cancer biomarkers and cancer progression Loss or absence of pro-u-plasminogen activator indicates that a cancer is high stage, high grade, or both Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of urine samples for bladder cancer |
Table 20.3
Publications of breast cancer biomarker studies indicating investigator interest in the field
Topic | Number of publications |
|---|---|
Biomarker | 593,113 |
Biomarker and cancer | 212,324 |
Biomarker and breast cancer | 27,988 |
Biomarker and breast cancer and genetics | 10,514 |
Biomarker and breast cancer and epigenetics | 45 |
Biomarker and breast cancer and methylation | 381 |
Biomarker and breast cancer and histone | 247 |
Biomarker and breast cancer and microRNA | 209 |
Biomarker and breast cancer and proteomics | 419 |
Biomarker and breast cancer and imaging | 1,094 |
Conclusion and Future Perspective
Considerable knowledge has been gained in understanding the biology of breast cancer and identifying biomarkers that can be used in detecting breast cancer, but translation of that knowledge to the clinical setting has been challenging [1]. Clinical validation is the main hurdle in the process. In a case-control study of the Prospect-EPIC (European Prospective Investigation into Cancer and Nutrition) study in which more than 300 breast cancer patients and matched controls were tested for breast cancer over a period of 3 years using a panel of eight serum markers (osteopontin, haptoglobin, cancer antigen 15-3, carcinoembryonic antigen, cancer antigen-125, prolactin, cancer antigen 19-9, and alpha-fetoprotein), very low specificity (50 %) and sensitivity (50 %) were observed [112]. This may be due to the presence of different breast cancer subtypes in collected samples. Such epidemiologic studies should select a broader target set of potential biomarkers that could be enabled by antibody array technologies in which profiles of up to 100 antibodies can be followed simultaneously. Making different groupings based on the status of hormone receptors (estrogen and progesterone) also might be helpful.
The need to identify and characterize early cancer diagnostic biomarkers is considerable because cancer is a heterogeneous disease, and a patient’s individual molecular profile, resulting from tumor microenvironment, determines disease development and response to treatment [37, 113]. Tumor microenvironment is affected by several factors, including epigenetic factors of the cell. As noted above, a number of noninvasive biomarkers for early detection of breast cancers have been identified.
In the field of cancer prevention, the National Cancer Institute (NCI) has an ongoing study, “the Study of Tamoxifen and Raloxifene (STAR),” with the objective of examining how the drug raloxifene compares with the drug tamoxifen in reducing the incidence of breast cancer in postmenopausal women who are at increased risk of developing the disease. Early results indicated that both drugs are equally effective and that about one-half of the disease incidence could be reduced. After the trial was continued for a longer time in the absence of these drugs to see whether previous prevention could be sustained, a 50 % reduction in incidence was seen with raloxifene, while a 38 % reduction was seen with tamoxifen. Both noninvasive and invasive cancers were studied in the STAR. Furthermore, participants taking raloxifene showed fewer side effects (e.g., uterine cancer, blood clots, cataracts) than those taking tamoxifen. Researchers with the National Surgical Adjuvant Breast and Bowel Project (NSABP) are conducting this study (financially supported by the NCI, USA), and more than 500 participants aged 35 years or older are involved in the study. The trial began in 1999 and stopped enrolling new patients in 2004.
The main areas in which progress and attention are needed with respect to biomarker identification are cost, high throughput, and the application of breast cancer biomarkers in clinical settings. Proper analytical and clinical validation of early markers has not been achieved. Currently, the key challenge in the field is the clinical validation of identified biomarkers. The NCI has developed guidelines for the analytical and clinical validation of biomarkers, but no biomarkers have been validated to date [37, 113–117]. Integrating genomic and proteomic markers with epigenetic markers may be useful in subtyping different cancers and cancer stages [46, 118]. Methylation profiling results from blood and tissues often differ. Koestler and colleagues conducted a systematic epigenome-wide methylation analysis and demonstrated that shifts in leukocyte subpopulations might account for a considerable proportion of variability in these patterns [119]. Multiplexing of biomarkers may reduce false-positive results in screening studies when the intent is to identify populations who are at high risk of developing breast cancer.
Quantitative imaging data storage and maintenance present their own challenges, as discussed above. Whether miR expression is localized in a specific part of the breast tissue must be evaluated carefully. In a tissue biopsy, the local concentration (number of miRs) may be low or high. Determining the accurate level of miR concentration is critical.
Association studies are extremely powerful in identifying new low-penetrance SNPs (biomarkers) that may have therapeutic implications. Identifying common low-susceptibility alleles is useful because it provides possible insight into the mechanisms of tumor biology and identification of high-risk individuals. Because genotyping is no longer expensive, the information from such studies can be utilized in personalized medicine in the form of targeted primary and secondary prevention.
Prognostication is a promising area in the translation of experimental research into clinical practice. In this approach, patterns of altered gene expression in tumors are used to construct classifiers instead of standard indices such as Nottingham Prognostic Index, Adjuvant! Online, and PREDICT [120–122].
In conclusion, we would like to emphasize that considerable progress has been made in identifying breast cancer biomarkers that can be used in the complete spectrum of carcinogenesis, from risk assessment to survival follow-up. The information discussed in this chapter may be useful in developing new interventions and therapeutic targets.
Acknowledgments
I am thankful to Joanne Brodsky of SCG, Inc., for reading the manuscript and providing her suggestions.
References
1.
Tang SS, Gui GP. Biomarkers in the diagnosis of primary and recurrent breast cancer. Biomark Med. 2012;6(5):567–85.PubMed
2.
Ouyang G, Chen Y, Setkova L, Pawliszyn J. Calibration of solid-phase micro-extraction for quantitative analysis by gas chromatography. J Chromatogr A. 2005;1097(1–2):9–16.PubMed
3.
Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800.PubMed
4.
Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107(15):6994–9.PubMedPubMedCentral
5.
Desmedt C, Sotiriou C, Piccart-Gebhart MJ. Development and validation of gene expression profile signatures in early-stage breast cancer. Cancer Invest. 2009;27(1):1–10.PubMed
6.
Lee SC, Xu X, Chng WJ, Watson M, Lim YW, Wong CI, et al. Post-treatment tumor gene expression signatures are more predictive of treatment outcomes than baseline signatures in breast cancer. Pharmacogenet Genomics. 2009;19(11):833–42.PubMed
7.
Normanno N, De Luca A, Carotenuto P, Lamura L, Oliva I, D’Alessio A. Prognostic applications of gene expression signatures in breast cancer. Oncology. 2009;77 Suppl 1:2–8.PubMed
8.
Tebbit CL, Zhai J, Untch BR, Ellis MJ, Dressman HK, Bentley RC, et al. Novel tumor sampling strategies to enable microarray gene expression signatures in breast cancer: a study to determine feasibility and reproducibility in the context of clinical care. Breast Cancer Res Treat. 2009;118(3):635–43.PubMedPubMedCentral
9.
Boeri L, Canzonieri C, Cagioni C, Ornati F, Danesino C. Breast cancer and genetics. J Ultrasound. 2011;14(4):171–6.PubMedPubMedCentral
10.
Kirk R. Genetics: the crystal ball clears for breast cancer therapy? Nat Rev Clin Oncol. 2011;8(7):383.PubMed
11.
Lindeman GJ, Visvader JE. Hereditary breast cancer genetics–from clinical curiosities to mainstream paradigms. J Mammary Gland Biol Neoplasia. 2011;16(1):1–2.PubMed
12.
Hall P. Current knowledge and tomorrows challenges of breast, ovarian and prostate cancer genetics. J Intern Med. 2012;271(4):318–20.PubMed
13.
Gayther SA, Song H, Ramus SJ, Kjaer SK, Whittemore AS, Quaye L, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67(7):3027–35.PubMed
14.
Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82(4):937–48.PubMedPubMedCentral
15.
Pharoah PD, Caldas C. Genetics: how to validate a breast cancer prognostic signature. Nat Rev Clin Oncol. 2010;7(11):615–6.PubMed
16.
Jacobson DR, Fishman CL, Mills NE. Molecular genetic tumor markers in the early diagnosis and screening of non-small-cell lung cancer. Ann Oncol. 1995;6 Suppl 3:S3–8.PubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree