Fig. 16.1
Radiotherapy treatment involves complex interaction of physical, biological, and clinical factors. The successful informatics approach should be able to resolve this interaction “puzzle” in the observed treatment outcome (e.g., local control or toxicity) for each individual patient [21]
In this chapter, we provide an overview of the current status of data-driven outcome modeling techniques for predicting tumor response and normal tissue toxicities for patients who receive radiation treatment with special focus on the emerging role of machine learning approaches to improve outcome modeling and response prediction. Then, we present examples of radiotherapy data and its big data notion. Finally, we discuss the potentials and challenging obstacles to applying bioinformatics and machine learning strategies to radiotherapy outcome modeling.
16.2 Data-Driven Outcome Modeling
Radiotherapy outcome models could be divided according to the underlying principle into (1) analytical models, which employ biophysical understanding of irradiation effects such as the linear quadratic (LQ) model, and (2) data-driven models, which are phenomenological models and depend on parameters available from the collected clinical and dosimetric data [20]. In the context of data-driven and multivariable modeling of outcomes, the observed treatment outcome (e.g., TCP or NTCP) is considered as the result of mathematical mapping of several dosimetric, clinical, or biological input variables [19]. Mathematically this is expressed as: 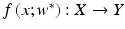 where R N (an input variable vector of N dimensions) is composed of the input metrics (dose-volume metrics, patient disease specific prognostic factors, or biological markers). The expression
where R N (an input variable vector of N dimensions) is composed of the input metrics (dose-volume metrics, patient disease specific prognostic factors, or biological markers). The expression 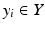 is the corresponding observed treatment outcome scalar. The variable w* includes the optimal parameters of model f(·) obtained by optimizing a certain objective functional. Learning is defined in this context as estimating dependencies from data [22]. The two common types of learning could be applied: supervised and unsupervised. Supervised learning is used when the endpoints of the treatments such as tumor control or toxicity grade are known; these endpoints are provided by experienced oncologists following RTOG or NCI criteria, and it is the most commonly used learning method in outcome modeling. Nevertheless, unsupervised methods such as principal component analysis (PCA) are also used to reduce dimensionality and to aid visualization of multivariate data and selection of learning method parameters [23]. The selection of the functional form of the model f(·) is closely related to the prior knowledge of the problem. In analytical models, the shape of the functional form is selected based on the clinical or biological process at hand; however, in data-driven models, the objective is usually to find a functional form that fits the data [24].
is the corresponding observed treatment outcome scalar. The variable w* includes the optimal parameters of model f(·) obtained by optimizing a certain objective functional. Learning is defined in this context as estimating dependencies from data [22]. The two common types of learning could be applied: supervised and unsupervised. Supervised learning is used when the endpoints of the treatments such as tumor control or toxicity grade are known; these endpoints are provided by experienced oncologists following RTOG or NCI criteria, and it is the most commonly used learning method in outcome modeling. Nevertheless, unsupervised methods such as principal component analysis (PCA) are also used to reduce dimensionality and to aid visualization of multivariate data and selection of learning method parameters [23]. The selection of the functional form of the model f(·) is closely related to the prior knowledge of the problem. In analytical models, the shape of the functional form is selected based on the clinical or biological process at hand; however, in data-driven models, the objective is usually to find a functional form that fits the data [24].
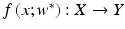 where R N (an input variable vector of N dimensions) is composed of the input metrics (dose-volume metrics, patient disease specific prognostic factors, or biological markers). The expression
where R N (an input variable vector of N dimensions) is composed of the input metrics (dose-volume metrics, patient disease specific prognostic factors, or biological markers). The expression 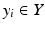 is the corresponding observed treatment outcome scalar. The variable w* includes the optimal parameters of model f(·) obtained by optimizing a certain objective functional. Learning is defined in this context as estimating dependencies from data [22]. The two common types of learning could be applied: supervised and unsupervised. Supervised learning is used when the endpoints of the treatments such as tumor control or toxicity grade are known; these endpoints are provided by experienced oncologists following RTOG or NCI criteria, and it is the most commonly used learning method in outcome modeling. Nevertheless, unsupervised methods such as principal component analysis (PCA) are also used to reduce dimensionality and to aid visualization of multivariate data and selection of learning method parameters [23]. The selection of the functional form of the model f(·) is closely related to the prior knowledge of the problem. In analytical models, the shape of the functional form is selected based on the clinical or biological process at hand; however, in data-driven models, the objective is usually to find a functional form that fits the data [24].
is the corresponding observed treatment outcome scalar. The variable w* includes the optimal parameters of model f(·) obtained by optimizing a certain objective functional. Learning is defined in this context as estimating dependencies from data [22]. The two common types of learning could be applied: supervised and unsupervised. Supervised learning is used when the endpoints of the treatments such as tumor control or toxicity grade are known; these endpoints are provided by experienced oncologists following RTOG or NCI criteria, and it is the most commonly used learning method in outcome modeling. Nevertheless, unsupervised methods such as principal component analysis (PCA) are also used to reduce dimensionality and to aid visualization of multivariate data and selection of learning method parameters [23]. The selection of the functional form of the model f(·) is closely related to the prior knowledge of the problem. In analytical models, the shape of the functional form is selected based on the clinical or biological process at hand; however, in data-driven models, the objective is usually to find a functional form that fits the data [24].16.3 Radiotherapy as a Big Data Resource
A typical radiotherapy treatment scenario can generate a large pool of “big data” that comprise but are not limited to patient demographics, volumetric dosimetric data about radiation exposure to the tumor and surrounding tissues, and 3D and 4D anatomical and functional disease longitudinal imaging features (radiomics), in addition to genomics and proteomics data derived from peripheral blood and tissue specimens. Accordingly, big data in radiotherapy could be divided based on its nature into four categories: clinical, dosimetric, imaging, and biological. These four categories of radiotherapy big data are described in the following.
16.3.1 Clinical Data
Clinical data in radiotherapy typically refers to cancer diagnostic information (site, histology, stage, grade, etc.) and patient-related characteristics (age, gender, comorbidities, etc.). In some instances, other treatment modalities information (surgery, chemotherapy, hormonal treatment, etc.) would be also classified under this category. The mining of such data could be challenging if the data is unstructured; however, there are good opportunities for natural language processing (NLP) techniques to assist in the organization of data [25].
16.3.2 Dosimetric Data
This type of data is related to the treatment planning process in radiotherapy, which involves radiation dose simulation using computed tomography imaging, specifically dose-volume metrics derived from dose-volume histograms (DVHs) graphs. Dose-volume metrics have been extensively studied in the radiation oncology literature for outcome modeling [14–17, 26, 27]. These metrics are extracted from the DVH such as volume receiving certain dose (Vx); minimum dose to x% volume (D x ); mean, maximum, and minimum dose; etc. More details are in our review chapter [20]. Moreover, we have developed a dedicated software tool called “Dose response explorer” (DREES) for deriving these metrics and modeling of radiotherapy response [28].
16.3.3 Radiomics (Imaging Features)
kV x-ray computed tomography (kV-CT) has been historically considered the standard modality for treatment planning in radiotherapy because of its ability to provide electron density information for target definition, structures, and heterogeneous dose calculations [2, 29]. However, additional information from other imaging modalities could be used to improve treatment monitoring and prognosis in different cancer types. For example, physiological information (tumor metabolism, proliferation, necrosis, hypoxic regions, etc.) can be collected directly from nuclear imaging modalities such as SPECT and PET or indirectly from MRI [30, 31]. The complementary nature of these different imaging modalities has led to efforts toward combining information to achieve better treatment outcomes. For instance, PET/CT has been utilized for staging, planning, and assessment of response to radiation therapy [32, 33]. Similarly, MRI has been applied in tumor delineation and assessing toxicities in head and neck cancers [34, 35]. Moreover, quantitative information from hybrid-imaging modalities could be related to biological and clinical endpoints, a new emerging field referred to as “radiomics” [36, 37]. In our previous work, we demonstrated the potential of this new field to monitor and predict response to radiotherapy in head and neck [38], cervix [38, 39], and lung [40] cancers, in turn allowing for adapting and individualizing treatment.
16.3.4 Biological Markers
A biomarker is defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathological processes, or pharmacological responses to a therapeutic intervention” [41]. Biomarkers can be categorized based on the biochemical source of the marker into exogenous or endogenous. Exogenous biomarkers are based on introducing a foreign substance into the patient’s body such as those used in molecular imaging as discussed above. Conversely, endogenous biomarkers can further be classified as (1) “expression biomarkers,” measuring changes in gene expression or protein levels, or (2) “genetic biomarkers,” based on variations, for tumors or normal tissues, in the underlying DNA genetic code. Measurements are typically based on tissue or fluid specimens, which are analyzed using molecular biology laboratory techniques [42]. Expression biomarkers are the result of gene expression changes in tissues or bodily fluids due to the disease or normal tissues’ response to treatment [43]. These biomarkers can be further divided into single parameter (e.g., prostate-specific antigen (PSA) levels in blood serum) versus bio-arrays. These can be based on disease pathophysiology or pharmacogenetic studies or they can be extracted from several methods, such as high-throughput gene expression (aka transcriptomics) [44–46], resulting protein expressions (aka proteomics) [47, 48], or metabolites (aka metabolomics) [49, 50]. On the other hand, the inherent genetic variability of the human genome is an emerging resource for studying disposition to cancer and the variability of patient responses to therapeutic agents. These variations in the DNA sequences of humans, in particular single-nucleotide polymorphisms (SNPs), have strong potential to elucidate complex disease onset and response in cancer [51]. Methods based on the candidate gene approach and high throughput (genome-wide associations (GWAS) studies) are currently heavily investigated to analyze the functional effect of SNPs in predicting response to radiotherapy [52–54]. There are several ongoing SNP genotyping initiatives in radiation oncology, including the pan-European GENEPI project [55], the British RAPPER project [56], the Japanese RadGenomics project [57], and the US Gene-PARE project [58]. An international consortium has been also established to coordinate and lead efforts in this area [59]. Examples include the identification of SNPs related to radiation toxicity in prostate cancer treatment [60–62].
16.4 Systems Radiobiology
To integrate heterogeneous big data in radiotherapy, engineering-inspired system approaches would have great potential to achieve this goal. Systems biology has emerged as a new field to apply systematic study of complex interactions to biological systems [63], but its application to radiation oncology, despite this potential, has been unfortunately limited to date [64, 65]. Recently, Eschrich et al. presented systems biology approach for identifying biomarkers related to radiosensitivity in different cancer cell lines using linear regression to correlate gene expression with survival fraction measurements [66]. However, such a linear regression model may lack the ability to account for higher-order interactions among the different genes and neglect the expected hierarchal relationships in signaling transduction of highly complex radiation response. It has been noted in the literature that modeling of molecular interactions could be represented using graphs of network connections as in power line grids. In this case, radiobiological data can be represented as a graph (network) where the nodes represent genes or proteins and the edges may represent similarities or interactions between these nodes. We have utilized such approach based on Bayesian networks for modeling dosimetric radiation pneumonitis relationships [67] and more recently in predicting local control from biological and dosimetric data [68].
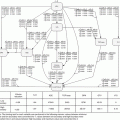
In the more general realm of bioinformatics, this systems approach could be represented as a part of a feedback treatment planning system as shown in Fig. 16.2, in which bioinformatics understanding of heterogeneous variables interactions could be used as an adaptive learning process to improve outcome modeling and personalization of radiotherapy regimens.
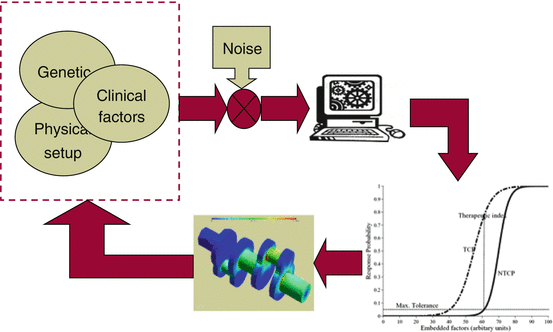

Fig. 16.2
The bioinformatics understanding of heterogeneous variable interactions as a feedback into the treatment planning system to improve patient’s outcomes. A heterogeneous list of variables with their noisy characteristics are acquired from retrospective or prospective studies and fed into in a learning algorithm to derive estimates of TCP/NTCP, which is typically corrected based on feedback of newly tested patients or scientific and clinical findings
16.5 Software Tools for Outcome Modeling
Many of the TCP/NTCP outcome modeling methods require dedicated software tools for implementation. Examples of such software tools in the literature are BIOPLAN and DREES. BIOPLAN (BIOlogical evaluation of treatment PLANs) uses several analytical models for evaluation of radiotherapy treatment plans [69], while DREES is an open-source software package developed by our group for dose-response modeling using analytical and data-driven methods [28] presented in Fig. 16.3. It should be mentioned that several commercial treatment planning systems have currently incorporated different TCP/NTCP models, mainly analytical ones that could be used for ranking and biological optimization purposes. A discussion of these models and their quality assurance guidelines is provided in TG-166 [11].
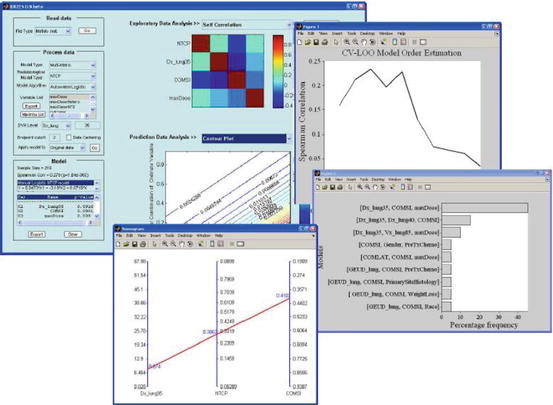

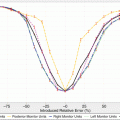
Fig. 16.3
DREES allows for TCP/NTCP analytical and multivariate modeling of outcomes data. The example is for lung injury. The components shown here are Main GUI, model order and parameter selection by resampling methods, and a nomogram of outcome as function of mean dose and location
16.6 Discussion
16.6.1 Data Sharing
Successful outcome modeling requires large datasets to meet statistical requirements, and sharing data is necessary to achieve this purpose. However, data sharing remains an issue for nontechnical issues [70]. Therefore, the Quantitative Analyses of Normal Tissue Effects in the Clinic (QUANTEC) consortium has suggested that cooperative groups adopt a policy of anonymizing clinical trial data and making these data publicly accessible after a reasonable delay. This delay would enable publication of all the investigator-driven, planned studies while encouraging the establishment of key databanks of linked treatment planning, imaging, and outcomes data [71]. An alternative approach is to apply rapid learning as suggested by the Maastro clinic group at Maastricht, in which innovative information technologies are developed that support semantic interoperability and enable distributed learning and data sharing without the need for the data to leave the hospital or the institution [72]. An example of multi-institutional data sharing is developed by the groups of Maastro clinic and the Policlinico Universitario Agostino Gemelli in Rome, Italy (Gemelli) [73].
16.6.2 Lack of Web Resources for Radiobiology
As of today, there are no dedicated web resources for bioinformatics studies in radiation oncology. Nevertheless, radiotherapy biological marker studies can still benefit from existing bioinformatics resources for pharmacogenomic studies that contain databases and tools for genomic, proteomic, and functional analysis as reviewed by Yan [74]. For example, the National Center for Biotechnology Information (NCBI) site hosts databases such as GenBank, dbSNP, Online Mendelian Inheritance in Man (OMIM), and genetic search tools such as BLAST. In addition, the Protein Data Bank (PDB) and the program CPHmodels are useful for protein structure three-dimensional modeling. The Human Genome Variation Database (HGVbase) contains information on physical and functional relationships between sequence variations and neighboring genes. Pattern analysis using PROSITE and Pfam databases can help correlate sequence structures to functional motifs such as phosphorylation [74]. Biological pathway construction and analysis is an emerging field in computational biology that aims to bridge the gap between biomarker findings in clinical studies with underlying biological processes. Several public databases and tools are being established for annotating and storing known pathways such as KEGG and Reactome projects or commercial ones such as the IPA or MetaCore [75]. Statistical tools are used to properly map data from gene/protein differential experiments into the different pathways such as mixed effect models [76] or enrichment analysis [77].
16.6.3 Protecting the Confidentiality and Privacy of Clinical Phenotype Data
QUANTEC offered a solution to radiotherapy digital data (treatment planning, imaging, and outcomes data) accessibility by asking cooperative groups to adopt a policy of anonymizing clinical trial data and making the data publicly accessible after a reasonable delay [71]. With regard to blood or tissue samples, no recommendation was made, however, by extending the same work and making any gene or protein expression assay measurements available under the same umbrella, while raw specimen data could be accessed from the biospecimen resource. For example, in the RTOG biospecimen standard operating procedure (SOP), it is highlighted that biospecimens received by the RTOG Biospecimen Resource are de-identified of all patient health identifiers and are enrolled in an approved RTOG study. Each patient being enrolled by an institution has to qualify and consent to be part of the study before being assigned a case and study ID by the RTOG Statistical Center. No information containing specific patient health identifiers is maintained by the Resource Freezerworks database, which is primarily an inventory and tracking system. In addition, information related to medical identifiers and any code lists could be removed completely from the dataset after a certain period say 10 years or so. Moreover, it has been argued that current measures by the Health Insurance Portability and Accountability Act (HIPPA) of 18 data elements are not sufficient and techniques based on research in privacy-preserving data mining, disclosure risk assessment data de-identification, obfuscation, and protection may need to be adopted to achieve better protection of confidentiality [78].
16.7 Future Research Directions
The ability to maintain high-fidelity large-scale data for radiotherapy studies remains a major challenge despite the high volume of clinical generated data on almost daily basis. As discussed above there have been several ongoing institutional and multi-institutional initiatives such as the RTOG, radiogenomics consortium, and EuroCAT to develop such infrastructure; however, there is plenty of work to be done to overcome issues related to, data sharing hurdles, patient confidentiality issues lack of signaling pathways databases of radiation response, development of cost-effective multicenter communication systems that allows transmission, storage, and query of large datasets such images, dosimetry, and biomarkers information. The use of NLP techniques is a promising approach in organizing unstructured clinical data. Dosimetry and imaging data can benefit from existing infrastructure for Picture Archiving and Communication Systems (PACS) or other medical image databases. Methods based on the new emerging field of systems radiobiology will continue to grow on a rapid pace, but they could also benefit immensely from the development of specialized radiation response signaling pathway databases analogous to the currently existing pharmacogenomics databases. Data sharing among different institutions is a major hurdle, which could be solved through cooperative groups or distributed databases by developing in a cost-effective manner the necessary bioinformatics and communication infrastructure using open-access resources through partnership with industry.
16.8 Conclusion
Recent evolution in radiotherapy imaging and biotechnology has generated enormous amount of big data that spans clinical, dosimetric, imaging, and biological markers. This data provided new opportunities for reshaping our understanding of radiotherapy response and outcome modeling. However, the complexity of this data and the variability of tumor and normal tissue responses would render the utilization of advanced bioinformatics and machine learning methods as indispensible tools for better delineation of radiation complex interaction mechanisms and basically a cornerstone to “making data dreams come true” [79]. However, it also posed new challenges for data aggregation, sharing, confidentiality, and analysis. Moreover, radiotherapy data constitutes a unique interface between physics and biology that can benefit from the general advances in biomedical informatics research such as systems biology and available web resources while still requiring the development of its own technologies to address specific issues related to this interface. Successful application and development of advanced data communication and bioinformatics tools for radiation oncology big data so to speak is essential to better predicting radiotherapy response to accompany other aforementioned technologies and usher significant progress toward the goal of personalized treatment planning and improving the quality of life for radiotherapy cancer patients.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree