Abstract
The virus infecting humans that is closest to SARS-CoV-2 is SARS-CoV, the agent that caused the SARS outbreak in 2002 and 2003. These two viruses are very similar in their genomes, in their way of entering cells, and in some of their clinical characteristics. Since 2003, we have learned many things from the virus that caused SARS. We have learned how the virus enters cells, how it replicates, and how it interacts with the immune system. We have learned some of the main factors that contribute to the worsening of the disease. Animal models have been established, and therapies have been developed and proposed. This acquired knowledge can accelerate the discovery of potential treatments for COVID-19.
When a disease insinuates itself so potently into the imagination of an era, it is often because it impinges on an anxiety latent within that imagination … SARS set off a panic about global spread and contagion at a time when globalism and social contagion were issues simmering nervously in the West.
The SARS outbreak provides evidence to support the hypothesis that southern China could be a site for emerging pandemic infectious disease in the future.
The virus infecting humans that is closest to SARS-CoV-2 is SARS-CoV, the agent that caused the SARS outbreak in 2002 and 2003. These two viruses are very similar in their genomes, in their way of entering cells, and in some of their clinical characteristics. Since 2003, we have learned many things from the virus that caused SARS. We have learned how the virus enters cells, how it replicates, and how it interacts with the immune system. We have learned some of the main factors that contribute to the worsening of the disease. Animal models have been established, and therapies have been developed and proposed. This acquired knowledge can accelerate the discovery of potential treatments for COVID-19.
There are many parallels that can be drawn between the outbreak of SARS in 2002–2003 and the outbreak of COVID-19 in 2019–2020. The two viruses likely started from the same pool of viruses, probably circulating in bats and infecting other species. Both outbreaks were identified as a cluster of pneumonias of unknown origin, and rapid response isolated and identified the viruses within two months. There are similarities in the clinical characteristics of the diseases, such as the incubation period, the mode of propagation, the first symptoms, and the high incidence of lower respiratory tract pneumonias. There are, however, intriguing differences. For instance, SARS was mostly localized in healthcare facilities and family clusters, whereas COVID-19 was able to spread easily beyond these settings. SARS was contained and practically disappeared within a few months, which is not the case for COVID-19. In this chapter, I examine some of these parallels and divergences.
How Did the SARS Outbreak Start?
In November 2002, the first case of an atypical pneumonia was reported in Foshan, near Guangzhou, in the Guangdong province in southern China. Several cases followed in the Guangdong region, including family clusters, suggesting an infectious spread. Many of the patients were transported to hospitals in Guangzhou, the Guangdong capital, for clinical care. This led to many infections within the hospitals there. The disease started with high fever, muscle pain, and respiratory symptoms (dry cough and shortness of breath), developing a few days later into pneumonia. When the WHO was informed in February 2003, the outbreak in Guangdong counted 305 cases and 5 deaths. The cluster included a large number of healthcare workers.
In February 2003, a doctor who treated SARS patients in Guangdong traveled to Hong Kong for a family gathering. He developed the disease while in Hong Kong and died there. Twenty-three other guests staying in the same hotel developed the disease. Many of the infected guests from the hotel traveled to other countries, including Canada, Taiwan, Singapore, and Vietnam, thus spreading the disease. By March 2003, SARS cases were identified in 13 countries, although most cases were reported in China and Hong Kong. The genomes of the viruses isolated in different regions were almost identical, suggesting that all cases were caused by the same agent that had emerged in a short time in the past. The disease was named severe acute respiratory syndrome, or SARS.
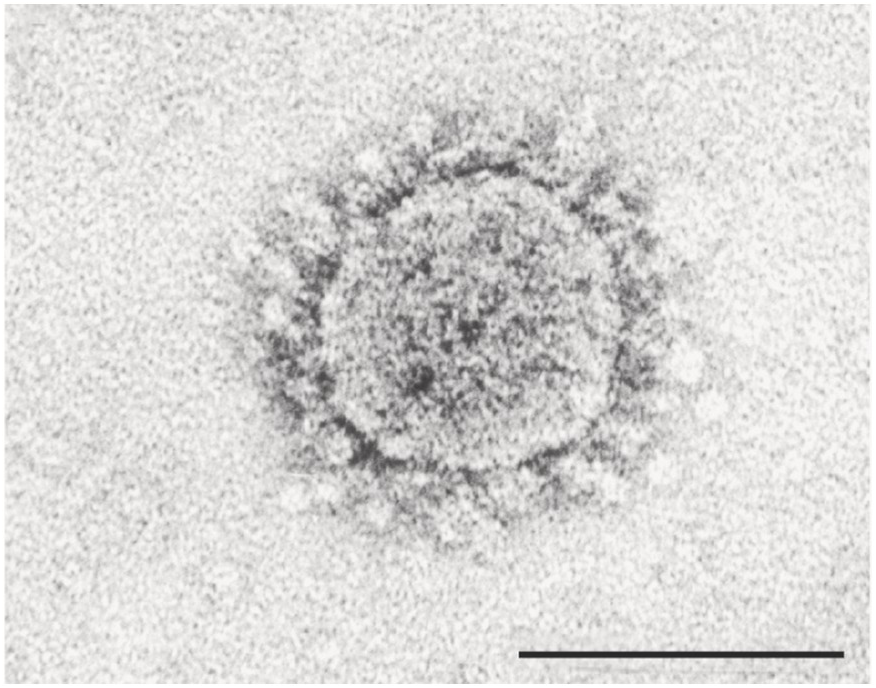
In April 2003, the agent causing SARS was isolated and was found to be a coronavirus distantly related to a previously known one (Figure 6.1). The virus was named SARS-CoV, as the SARS-causing coronavirus. None of the healthy controls showed antibody or viral RNA, suggesting that this was a new virus. It was also found that patients did not show immune response to the new virus, suggesting that the virus was new and not cross-reacting with the commonly circulating human coronaviruses 229E and OC43. All these analyses indicated that it was a new virus that the immune systems of the patients had not encountered before.
Figure 6.1 Identification of the virus causing SARS in 2003. Electron microscopy image of a SARS-CoV virus showing the typical coronavirus structure with spike projections surrounding the membrane envelope of the virus. The bar on the bottom right-hand side represents a scale of 0.00001 cm (100 nm). The virus was isolated four months after the first few cases were reported in Guangdong in 2002. The genome of the SARS-CoV virus was compared to previously known viral genomes and was found to be a distant relative of other coronaviruses. Very little was known at the time about betacoronaviruses circulating in other species. By contrast, during the 2019 SARS-CoV-2 outbreak, the time taken for the isolation and sequencing of the viral genome was reduced to less than a month after the first cases of the outbreak were reported. This quick action led to the implementation of detection kits and active surveillance and quarantine measures. The outbreak in Wuhan was effectively controlled by March 2020.
Where Was the SARS Virus Coming From?
The SARS virus was not circulating in humans, which triggered a search for the virus in other species. One potential source might be related to some culinary habits in the south of China, where wild game and live animal markets (wet markets) are not uncommon. This hypothesis was reinforced by the fact that some of the early SARS cases in Guangdong included workers at restaurants that featured wild animals on the menu.
In 2003, different studies indicated that the viruses could be isolated from wild animals sold in wet markets in the Guangdong province of China. SARS-like coronaviruses were found in Himalayan palm civets (also called masked palm civets, with the scientific name Paguma larvata) and in a raccoon dog in a live-animal market in Shenzhen. Sequencing of the genomes of these viruses indicated that they were very closely related to SARS-CoV, the virus responsible for the SARS outbreak.
Two cases were identified as a waitress and a customer in a restaurant in Guangzhou that served palm civets and had the live animals in the entrance of the restaurant. These animals included palm civets that tested positive for SARS-like viruses. This small outbreak did not grow further. These findings suggest that live-animal markets could increase the transmission of zoonotic viruses (viruses transmitted from animals to humans) through some of the species sold there.
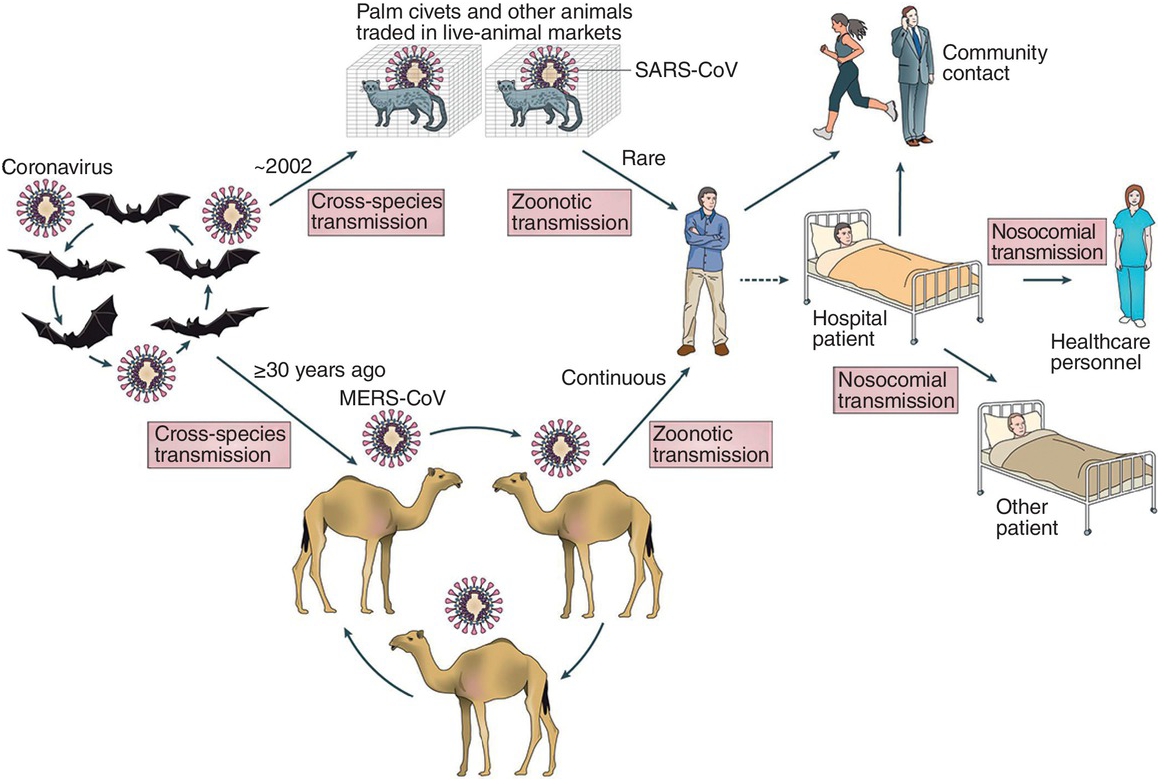
In 2004, further studies indicated that SARS-CoV-like viruses were circulating in various species of bats in different regions of China. Genomic characterization of these viruses indicated a close similarity to SARS-CoV and showed a large genetic diversity. Some bat species were widely found to have antibodies to SARS-like viruses, suggesting that bats could be the wildlife reservoir host for these viruses. In particular, horseshoe bats (Rhinolophus) were found to have large numbers of SARS-like coronaviruses. Further studies, published in 2017, found that viruses sampled from bats living in caves in Yunnan, in southwestern China, contained the different pieces of genomic material necessary for generating a SARS-CoV virus through a recombination process.
These findings suggest a possible scenario in which bat viruses can recombine and mutate to generate viruses able to infect humans (Figure 6.2). This could have happened through a mediating species, such as the palm civet. However, this is not necessary, as some bat SARS-CoV-like viruses are able to infect human cells, suggesting that a direct infection of SARS-CoV-like viruses is possible without any other mediating species.