MOLECULAR DIAGNOSTICS
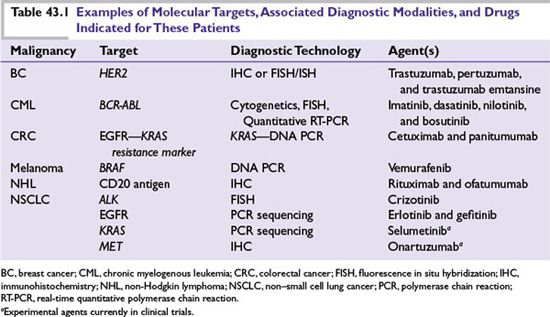
The more comprehensive understanding of the genetic makeup of tumors achieved in the past decade has resulted in molecular diagnoses being offered to patients with metastatic breast, colon, lung, central nervous system malignancies, melanoma, and leukemia/lymphoma. Diagnostic techniques commonly employed include immunohistochemistry (IHC), which can detect aberrant expression, localization, and phosphorylation levels of proteins; in situ hybridization (ISH), which reports on the localization and expression of messenger RNA (mRNA); fluorescence ISH (FISH), which can determine chromosomal abnormalities and gene copy number alterations; and polymerase chain reaction (PCR)-based methods, which can detect somatic mutations deletions or Fusions in key oncogenes in addition to quantifying mRNA expression. Formalin-fixed and paraffin-embedded (FFPE) tissues from surgical or biopsied specimens are often used for testing in the clinic. Other clinical specimens, such as fresh frozen tissues derived from biopsies, are also used to some extent, typically in the context of early drug development, to assess drug activity and mechanism of action, as well as to understand mechanisms of resistance upon progression. Table 43.1 provides examples of the types of molecular analyses that are used to inform the selection of therapeutic agents for patients.
In most of these examples, a single diagnostic test is used to identify specific “driver mutations” (genetic aberrations causally implicated in oncogenesis or tumor survival) for which targeted therapies exist. Validated, diagnostic assays that are approved by regulatory authorities to specifically guide treating physicians regarding which therapeutic option to choose are called “companion diagnostics.” Examples are diagnostic tests that detect the aberrant expression of specific therapeutic targets (e.g., human epidermal growth factor receptor 2 [HER2] in breast cancer) or factors predicting resistance to targeted agents (e.g., the presence of v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog [KRAS] in colorectal tumors that confer resistance to epidermal growth factor receptor [EGFR]-targeting antibodies). Although these represent significant advances, obtaining information on a single genetic variable from tumor samples at a single point in time may not be informative, as it does not take into consideration additional tumor, microenvironment, or host factors that may also affect treatment outcomes. Finally, the inevitable emergence of resistance to targeted therapies for the majority of patients with metastatic disease supports the need for the assessment of multiple molecular features using a variety of platforms serially over the course of the patient treatment journey.
Several techniques now allow for the simultaneous analysis of multiple genes in tumor tissue. Single nucleotide polymorphisms (SNPs) affecting metabolic enzymes in the genomes of individual patients have identified subsets of patients at increased risk of drug toxicity, resulting in now-routine screening and possible dose adjustments for those treated with thiopurines or irinotecan. SNPs arising from somatic mutations in tumor cells can now be analyzed in a high-throughput fashion using oligonucleotide probes bound to a solid support. Array-comparative genomic hybridization, examines DNA copy number changes and provides information on gene amplifications and deletions found in tumors. DNA microarrays are widely used in predictive and prognostic examinations of cancer genomes. These employ microscopic arrays of oligonucleotides or cDNA attached to solid supports (“gene chips”) to which fluorescently labeled DNA transcribed from sample RNA is hybridized, providing the data used to generate “gene signatures.” Although not yet practical for patient-level purposes, global genomic characterization of tumors using, in part, whole-genome DNA sequencing has been used to elucidate core signaling pathways in pancreatic cancer and glioblastoma multiforme (GBM), in the former case identifying a set of 12 core pathways and processes genetically altered in most tumors and in the latter case revealing a link between O6-methylguanine DNA-methyltransferase (MGMT) promoter methylation and a hypermutator phenotype in treated GBMs. Real-time quantitative PCR (RT-PCR) assays, although limited to the examinations of several hundred “candidate” genes rather than an entire genome, are increasingly used for tumor classification, in addition to predictive and prognostic purposes. Finally, direct analysis of proteins (“proteomics”) eliminates the potential discordance between mRNA and protein expression levels. Although more technically challenging to address in a high-throughput fashion than genomic methods, profiling of a limited number of proteins of interest in serum or tissue is now possible using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF).
RESISTANCE
Recent insights into the nature of innate and acquired resistance in cancer provides further support for the implementation of a multiplex diagnostics approach. As a comparison, these resistance mechanisms are similar to those found in infectious diseases, where they also represent a major challenge to therapy.
As highlighted in Table 43.2
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree