within the same species, exemplified by the large number of killer immunoglobulin superfamily (KIR) haplotypes found in humans.10 Studies of Ig superfamily (SF) genes expressed in the nervous system and immune system showed definitively that the immune system molecules evolve at a faster rate.11 Finally, rapid evolution of immune system molecules and mechanisms is the general rule, but molecules functioning at different levels of immune defense (recognition, signaling, or effector) can evolve at widely varying rates.
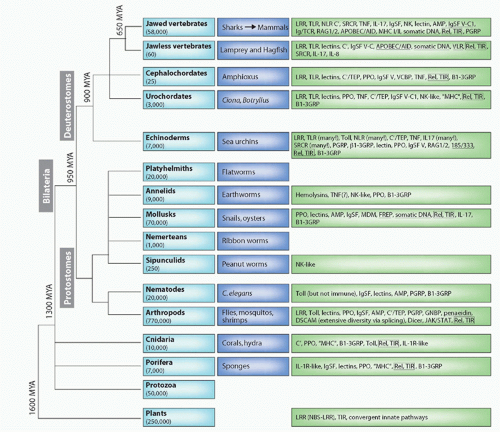 FIG. 4.1. Major Animal Groups and Immune Mechanisms/Molecules Described to Date in Each Group. The first box on the left in each row describes the animal taxon and the approximate number of species in that group. The next box shows specific examples of species or subgroups. The third box lists molecules/mechanisms found in each group: underlined terms indicate somatic changes to antigen receptors or secreted molecules. Figure modified from Hibino et al.48 and Flajnik and Du Pasquier.61 See Table 4.1 for definition of the acronyms. |
species (eg, the function[s] of γδ T cells). The “Janus paradigm,” therefore, can be quite useful when examining any pathway in the immune system.
determining the function of different molecules, best known for the Igs (transmembrane [TM] versus secreted forms, as well as inflammatory versus neutralizing forms in non-mammalian vertebrates). Further diversity can be obtained by the assembly of multichain receptors in which different components are combined. The classical example in the jawed vertebrate adaptive immune system is that of Ig light (L) and heavy (H) chains of antibodies but similar combination can occur with insect peptidoglycan-recognizing proteins (PGRP), vertebrate TLRs, and many others.
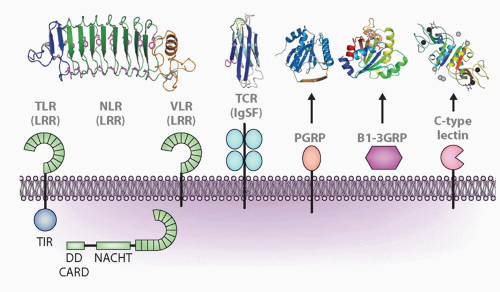 FIG. 4.2. Major Molecular Families Described in the Text and Representatives of Each Family: Leucine-Rich Repeats (LRRs), Immunoglobulin Superfamily (IgSF), Peptidoglycan-Recognition Protein (PGRP), β1-3 Glucan Recognition Protein (β1-3GRP), and C-Type Lectins. Representative structures are shown above for LRR (toll-like receptor, nucleotide-binding domain leucine-rich repeat [NLR], variable lymphocyte receptor), IgSF, PGRP, β1-3GRP, and C-type lectin. Other acronyms are defined in Table 4.1. For the NLR model, the echinoderms have N-terminal death domains, whereas all other animals have caspase-recruitment domains. Figure modified from Hibino et al.48 |
TABLE 4.1 Molecules and Abbreviations Found Throughout the Text | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
nervous system. The immunity role encompasses phagocytosis, encapsulation, and sometimes production of effector molecules (see Fig. 4.336). These roles all require recognition of pathogen-associated molecular patterns (PAMPs) or self-derived defense molecules (ie, opsonization) at the cell surface.37,38,39,40
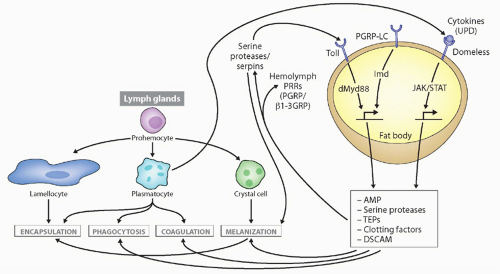 FIG. 4.3. Types of Immune Responses and Cells in Insects, with Drosophila as the Prototype. Secreted defense molecules are made by fat body cells in response to pathogens, which either act as direct effector molecules or feed back on hemocytes to stimulate their defense functions. Unpaired (UPD) is produced after virus infection, stimulating defense molecule upregulation via the JAK/signal transducer and activator of transcription pathway. Not shown is the RNAi pathway, also induced upon virus infection. Details on stimulation of the toll and immune deficiency pathways are found in Figure 4.4. This figure was modified from Lemaitre and Hoffmann.36 |
The hyaline cells are involved in the clotting process and the granular cells in phagocytosis, encapsulation, and the prophenoloxidase (PPO) pathway. The hematopoietic organ is located on the dorsal and dorsolateral regions of the stomach.39 Crustacean hemocytes can now be cultured and their response to virus can be examined,46 and markers of the three hematopoietic lineages are available.47
follows Srp expression, whereas in the lymph gland there is a long delay between Srp expression and the appearance of the lymph gland-derived hemocytes.38 Hematopoiesis in the head mesoderm and yolk sac may be related evolutionarily. A further similarity occurs at the AGM/lymph gland level in Drosophila. The lymph gland develops from a part of lateral mesoderm that also gives rise to vascular and excretory cells, much like the vertebrate AGM. The conserved relationship between blood precursors and vascular and excretory systems is intriguing.
phagocytes also leads to upregulation of the antigen processing machinery, costimulatory molecules, and proinflammatory cytokines that can enhance adaptive immunity.
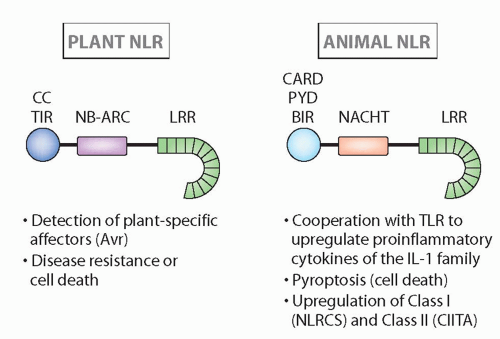 FIG. 4.4. Structure and Major Functions of Nucleotide-Binding Domain Leucine-Rich Repeats in Plants and Animals.14 Although the structures are quite similar, and recognition can be analogous, these families seem to have arisen via convergent evolution (see text for more details). |
clearly different from the ones expressed in neurons.24 In Drosophila, the DSCAM gene is composed of 115 exons, 95 of which encode alternative possibilities for splicing of exons 4, 6, and 9. The molecule consists of 10 IgSF domains and 6 FN domains, and present as either a membrane or soluble form, presumably generated by proteolysis of the membrane form. Each cell expresses only a fraction of the isoform repertoire.
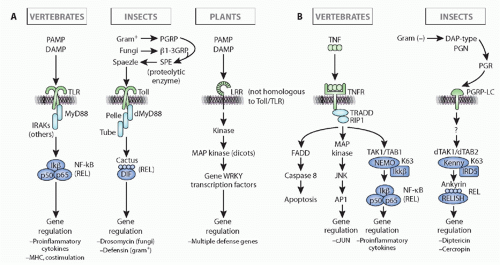 FIG. 4.5. Comparison of Immune Response Induction, Intracellular Pathways, and Immune Outcome in Insect (eg, Drosophila), Vertebrates (eg, human), and Plants (a Composite of Pathways in Monocots and Dicots). Note that the initiation of the response and the outcome(s) in insects and vertebrates are quite different for both the toll/toll-like receptor pathway (left, A) and the immune deficiency/tumor necrosis factor pathways (right, B), but the intracellular signaling pathways are well conserved evolutionarily in insect and vertebrate (details in the text). Note as well that plants use similar molecules for recognition (leucine-rich repeat-containing molecules) and have similar intracellular pathways with kinase cascades, but all of the molecules of recognition, signaling, and effector are derived by convergent evolution as compared to animals. This figure was modified from Beutler et al.152 |
expressed in many tissues such as gills, skin, and intestine, providing immune defense. They are expressed before the development of adaptive immunity and likely provide an important protective role.96 The human PGRP genes are found on the MHC chromosomal paralogs 1q21 and 19q13/p13. All detected splice-variant isoforms bind to bacteria and peptidoglycan. Like the fish molecules, mammalian PGRPs are also positioned at epithelial surfaces and promote intestinal homeostasis by discriminating somewhat between commensal (eg, lactobacilli) and pathogenic bacteria. Knockout mice have increased pathogenic bacteria on mucosal surfaces that induce colitis after injury in the dextran sulphate sodium autoimmune assay.97
heralded the molecular analyses of innate immunity in the invertebrates.106
the classical pathway of complement by splitting of C4 and C2 (the same function as C1s; MASP2 appears to be the active protease), but also can activate the alternative pathway in ways that are not understood and thus completely bypass the classical pathway. Indeed, MASP-1 and -2 are homologs of C1r and C1s (see Fig. 4.6). Both C1q and MBL can promote the uptake of apoptotic bodies by phagocytes, via collectin receptors. Another lectin, ficolin, can also initiate the MASP pathway,113 and it would not be surprising if other activators were discovered in the future (eg, the ancient molecule C-reactive protein is also capable of activating C′). Finally, the classical pathway, which is dependent upon antibody molecules bound to a surface, results in the same potential effector outcomes described previously for the alternative pathway. Novel molecules initiating this pathway are C1q, C1r, C1s, C4, and C2, as well as specific negative regulatory proteins such as C4-binding protein.
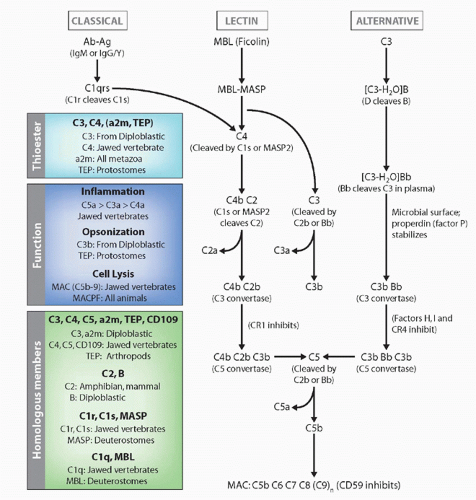 FIG. 4.6. Evolution of the Complement System. The general pathways and appearance of the various components in the phylogenetic tree are emphasized. |
as suggested by recent pioneering experiments in Ciona and Styela, it will be interesting to determine whether they are involved in some type of inflammation, thought to be the domain of the vertebrates.129
strategy to preserve this interaction; perhaps the CD94 homologs of invertebrates are genetically linked to genes encoding their ligands. In addition, as described in the following, the close genetic linkage of receptor and ligand genes is a common theme in “histocompatibility reactions” throughout the animal and plant kingdoms described below.
this view has changed (see the following). The secretory µ H chain is found in all vertebrates, usually consists of one V and four C1 domains, and is heavily glycosylated. H chains associate with each other and with L chains through disulfide bridges in most species, and IgM subunits form pentamers or hexamers in all vertebrate classes except teleost fish, which form tetramers.189 The µ CH4 domain is most evolutionarily conserved, especially in its C-terminal region, whereas the CH2 domain evolves at the fastest rate.188 There are several µ-specific residues in each of the four CH domains among vertebrates suggesting a continuous line of evolution, which is supported by phylogenetic analyses. Like TCR TM regions, µ TM regions are also well conserved among sharks, mammals, and amphibians, but the process by which the Ig TM messenger RNA is assembled varies in different species. In all vertebrate classes except teleosts, the µ TM region is encoded by separate exons that are spliced to a site on µ messenger RNA located approximately 30 basepairs from the end of the CH4-encoding exon. By contrast, splicing of teleost fish µ messenger RNA takes place at the end of CH3 exon.190 In holostean fish (gar and sturgeon), cryptic splice donor sites are found in the CH4 sequence that could lead to conventional splicing, but in the bowfin there is another cryptic splice donor site in CH3.191 The TM region itself is interesting as it is the only one that does not contain a residue capable of making an ionic bond with the ITAM-containing molecule (in this case, Ig-α and Ig-β.) As mentioned, some modifications apparently related to the particular environment were noticed in the Antarctic fish Trematomus bernacchii. There are two remarkable insertions, one at the VH-CH1 boundary and another at the CH2-CH3 boundary; the latter insertion results in a very long CH2-CH3 hinge region. Rates of nonsynonymous substitutions were high in the modified regions, suggesting strong selection for these modifications. These unusual features (also unique glycosylation sites) may permit flexibility of this IgM at very low temperatures.192
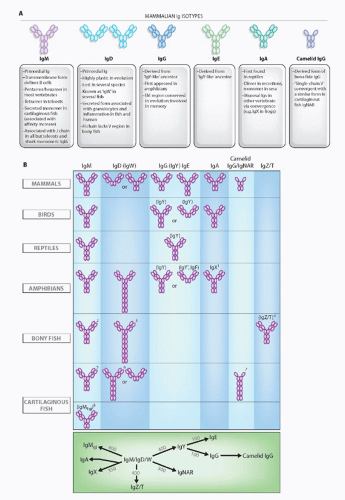 FIG. 4.7. Immunoglobulin (Ig) Isotypes in the Jawed Certebrates. A: Mammalian isotypes and their relationships to Igs in vertebrates from other classes. Each oval represents an Ig superfamily C1 domain. IgD is shown in two forms, mouse (left) and human (right).2 B: Major Ig isotypes in all vertebrates. The bottom panel displays the approximate divergence times of all isotypes. IgM/D/W was found at the inception of adaptive immunity. 1, IgX is in the IgA column because it is preferentially expressed in the intestine, and IgA seems to have been derived from an IgX ancestor; IgX seems to have been derived from both IgM and IgY ancestors; 2, secreted IgM in teleost fish is a tetramer, and the transmembrane (TM) form only has three C domains; 3, the teleost fish IgD H chains incorporate the µC1 domain via alternative splicing in the TM form, and the secretory form does not have a V region and does not associate with L chains; 4, the new bony fish isotype, IgZ/T, may not be found in all fish species; 5, the secreted form of shark/skate IgM is present as a pentamer and monomer at approximately equal levels; 6, the TM form of IgW has four C domains; 7, a major TM form of IgNAR has three C domains, and IgNAR is related to camelid IgG by convergent evolution; 8, no TM form has been found (to date) for IgM1gj. The bottom panel displays the approximate divergence times of all isotypes. IgM/D/W was found at the inception of adaptive immunity. |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








